Research Interests
Computational Nanoscale Imaging
We develop new methods for diffractive imaging of nanoscale objects, primarily using X-rays. Within this area, we work on developing analysis algorithms, often involving the solving of ill-posed inverse problems, and applying them to experimental data.
Biomolecular single particle imaging (SPI)
Using the power of XFEL pulses to image uncrystallized biomolecules
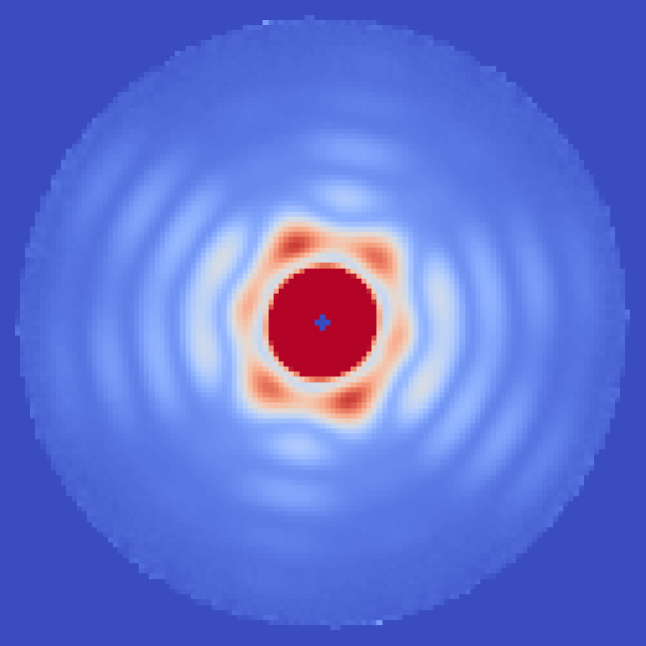
Coherent diffractive imaging (CDI) is a technique to determine the structure of an object from the far-field diffraction pattern produced when it interacts with a coherent wave. Among other things, one variation of this technique can be used to determine the 3D structure of biomolecules and other nanoscale objects using X-rays. Unfortunately, these small objects scatter too little to see much in a single image before radiation damage kicks in and destroys the particle.
X-ray free electron lasers (XFELs) have bright enough and short enough pulses that one can combine the information from a large number of molecules and generate a 3D structure by computationally determining the orientation and other unmeasured (latent) variables.1
Our group performs XFEL experiments which push the limits of these techniques2, and then execute the entire analysis pipeline from raw data to structure. Recently, we have been pursuing a holographic reference-enhancement configuration to improve the efficiency of imaging weakly scattering biomolecules.3
References:
1. Ayyer, Lan, et al. "Dragonfly: an implementation of the expand–maximize–compress algorithm for single‐particle imaging." J. Appl. Cryst. 49 (4), 1320-1335 (2016)
2. Ayyer, Xavier, et al. "3D diffractive imaging of nanoparticle ensembles using an X-ray laser." Optica 8(1), 15-23 (2021)
3. Ayyer, "Reference-enhanced x-ray single particle imaging. " Optica 7(6), 593-601 (2020)
Serial Diffractive Imaging to Understand Heterogeneous Ensembles
Combining SPI with machine learning to better image dynamics
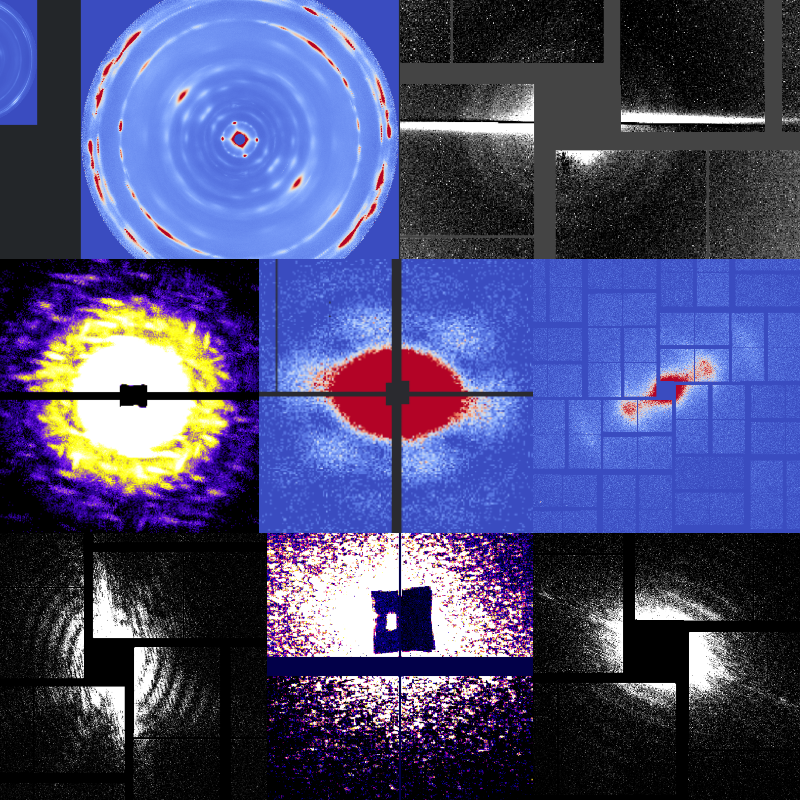
Diffractive imaging experiments often involve exposing thousands to millions of objects one at a time before using computational methods to assemble them into a single dataset.1
As an example, in X-ray SPI, patterns are oriented and merged to produce a single 3D intensity distribution. This requires all objects to be identical, upto orientation. But even when this is not the case, it is often interesting to understand the structural variations in such ensembles.2
Alternatively, we may want to track a reaction or phase transition which one cannot precisely trigger and classify the large number of patterns along the "reaction coordinate". In all these cases, machine learning methods such as deep learning using an appropriate feature set can be used to understand the dynamics beyond conventional ensemble-averaged measurements.
References:
1. Ayyer, Xavier, et al. "3D diffractive imaging of nanoparticle ensembles using an X-ray laser." Optica 8(1), 15-23 (2021)
2. Zhuang, et al. "Unsupervised learning approaches to characterize heterogeneous samples using X-ray single particle imaging." IUCrJ 9(2) (2022)
Protein crystal diffuse scattering
Extracting structural information and equilibrium dynamics from imperfect crystals
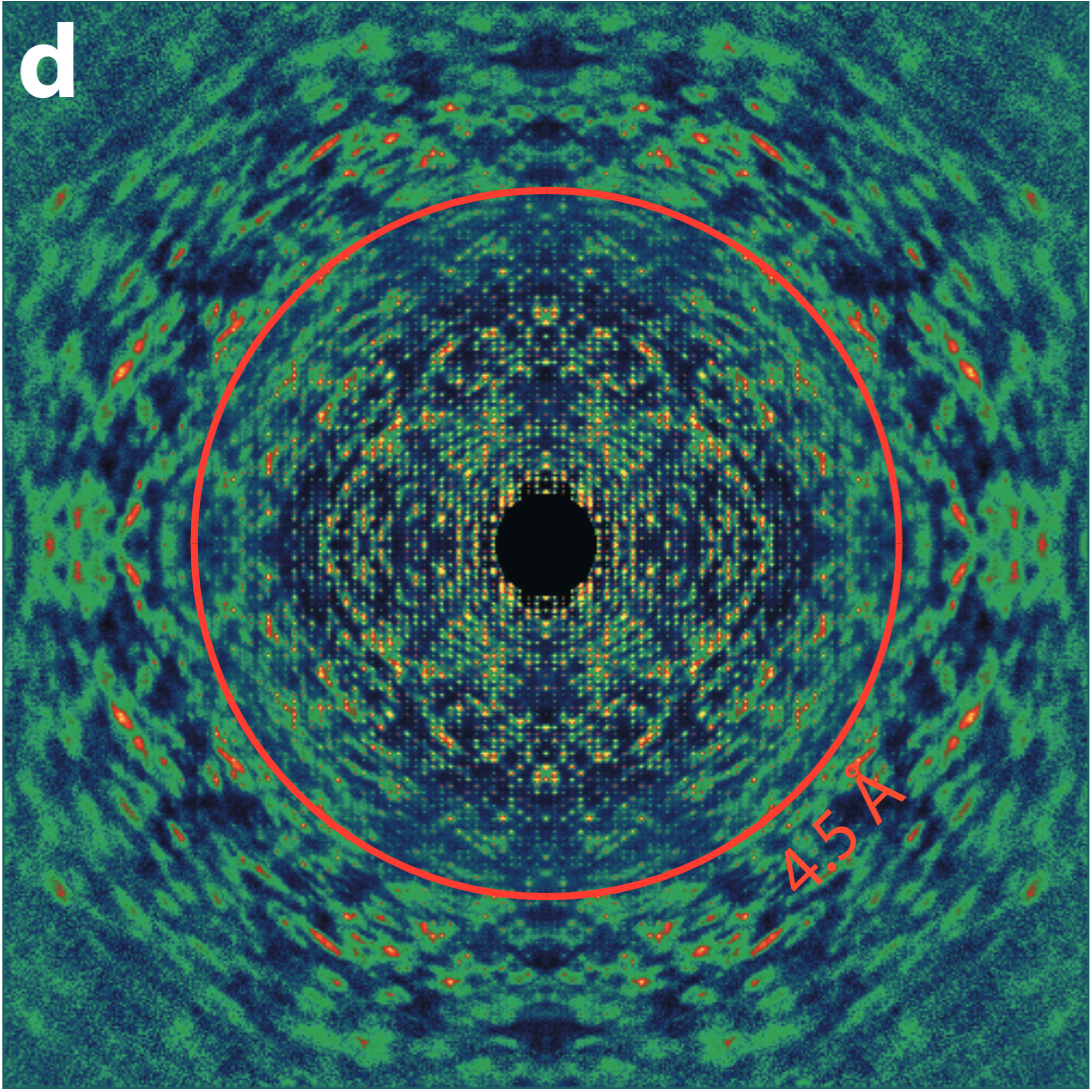
X-ray crystallography remains the dominant method to obtain high resolution structures of proteins and other biomolecules. This is because the diffraction signal is concentrated in bright Bragg peaks, giving good signal-to-noise to high resolution and making subtracting the diffuse background easier.
However, part of this diffuse 'background' is cause by the disorder in these crystals. And in fact, if the disorder is correlated i.e. multiple atoms moving coherently, the scattering pattern has features which inform us about this coherent motion1. Thus, we get information about how the proteins move naturally.
One component of disorder seen in some protein crystals is rigid-body-like motion where the entire molecule moves together, primarily because the crystal contacts are weaker than the internal bonding of the tertiary and quaternary structure. In these cases, the diffuse scattering is directly related to the molecular transform, and so can be used to get a higher resolution structural model of the protein2.
We are interested in refining this method of improving resolution in the case of rigid-body-like disorder, but also in developing methods to reliably extract more complex modes of motion from any protein crystal diffaction pattern.
References:
1. Ayyer, Chapman, Yefanov. "Structure Determination by Continuous Diffraction from Imperfect Crystals" In: X-ray Free Electron Lasers. Springer, Cham (2018)
2. Ayyer, Yefanov, et al. "Macromolecular Imaging with Imperfect Crystals", Nature 530 (7589), 202-206 (2016)
3. Mazumder, Ayyer. "Understanding conformational dynamics from macromolecular crystal diffuse scattering." bioRxiv 2021.02.11.429988 (2021)
Incoherent diffractive imaging (IDI)
Using 2-photon correlations to obtain structural insight from fluorescence
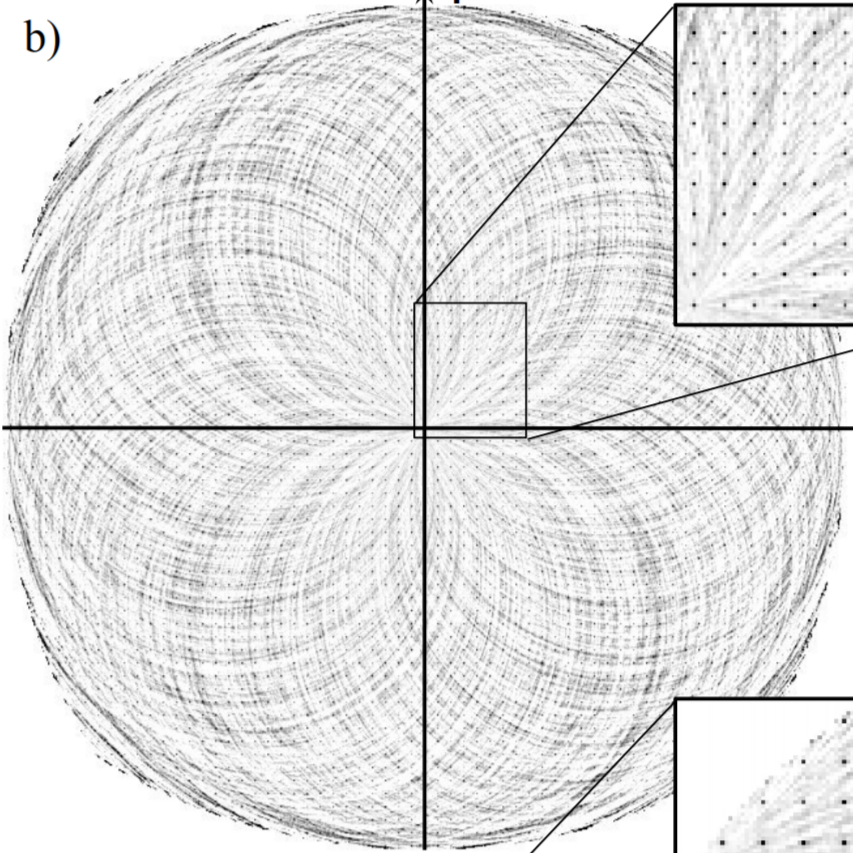
Incoherent light does not produce stable interference patterns (the overlapping light from two red LEDs won't create fringes). However, they do produce transient patterns which are stable over the coherence time. If we can measure these transient patterns and somehow average the information, we can obtain the structure of the source distribution.
The interference is not stable because each wave packet produced has a random initial phase. However, if we average the 2-point intensity correlation of each pattern, the random phases average out and we are left with the phase difference due to path difference (the structural part)1. This effect was first demonstrated in astronomy by Hanbury Brown and Twiss.
We have demonstrated this effect with atomic fluorescence2, which also has the same random phases and we want to develop this into an imaging technique. One powerful application of this technique would be sensitivity to the energy of the emitted photon. The degree of interference would depend on the electronic states of the interfering atoms, suggeting the tantalizing possibility of performing spectroscopy with atomic spatial resolution.
One can also combine this method with conventional SPI to study heterogeneous catalyst nanoparticles made of more than one atom type. Using orientation information from SPI, the IDI reconstruction gives us the substructure of each atom type independently.
References:
1. Classen, Ayyer, et al. "Incoherent Diffactive Imaging via Intensity Correlations of hard X-rays." Phys. Rev. Lett. 119 (5), 053401 (2017)
2. Trost, Ayyer, et al. "Imaging via Correlation of X-Ray Fluorescence Photons." Phys. Rev. Lett. 130, 173201 (2023)